PyBroom Example - Multiple Datasets - Minimize¶
This notebook is part of pybroom.
This notebook demonstrate using pybroom when performing Maximum-Likelihood fitting (scalar minimization as opposed to curve fitting) of a set of datasets with lmfit.minimize. We will show that pybroom greatly simplifies comparing, filtering and plotting fit results from multiple datasets. For an example using curve fitting see pybroom-example-multi-datasets.
In [1]:
%matplotlib inline
%config InlineBackend.figure_format='retina' # for hi-dpi displays
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.pylab import normpdf
import seaborn as sns
from lmfit import Model
import lmfit
print('lmfit: %s' % lmfit.__version__)
lmfit: 0.9.5
/home/docs/checkouts/readthedocs.org/user_builds/pybroom/envs/stable/lib/python3.4/site-packages/IPython/html.py:14: ShimWarning: The `IPython.html` package has been deprecated. You should import from `notebook` instead. `IPython.html.widgets` has moved to `ipywidgets`.
"`IPython.html.widgets` has moved to `ipywidgets`.", ShimWarning)
In [2]:
sns.set_style('whitegrid')
In [3]:
import pybroom as br
Create Noisy Data¶
Simulate N datasets which are identical except for the additive noise.
In [4]:
N = 20 # number of datasets
n = 1000 # number of sample in each dataset
np.random.seed(1)
d1 = np.random.randn(20, int(0.6*n))*0.5 - 2
d2 = np.random.randn(20, int(0.4*n))*1.5 + 2
d = np.hstack((d1, d2))
In [5]:
ds = pd.DataFrame(data=d, columns=range(d.shape[1])).stack().reset_index()
ds.columns = ['dataset', 'sample', 'data']
ds.head()
Out[5]:
| dataset | sample | data | |
|---|---|---|---|
| 0 | 0 | 0 | -1.187827 |
| 1 | 0 | 1 | -2.305878 |
| 2 | 0 | 2 | -2.264086 |
| 3 | 0 | 3 | -2.536484 |
| 4 | 0 | 4 | -1.567296 |
In [6]:
kws = dict(bins = np.arange(-5, 5.1, 0.1), histtype='step',
lw=2, color='k', alpha=0.1)
for i in range(N):
ds.loc[ds.dataset == i, :].data.plot.hist(**kws)
Model Fitting¶
Two-peaks model¶
Here, we use a Gaussian mixture distribution for fitting the data.
We fit the data using the Maximum-Likelihood method, i.e. we minimize the (negative) log-likelihood function:
In [7]:
# Model PDF to be maximized
def model_pdf(x, a2, mu1, mu2, sig1, sig2):
a1 = 1 - a2
return (a1 * normpdf(x, mu1, sig1) +
a2 * normpdf(x, mu2, sig2))
# Function to be minimized by lmfit
def log_likelihood_lmfit(params, x):
pnames = ('a2', 'mu1', 'mu2', 'sig1', 'sig2')
kws = {n: params[n] for n in pnames}
return -np.log(model_pdf(x, **kws)).sum()
We define the parameters and “fit” the \(N\) datasets by minimizing
the (scalar) function log_likelihood_lmfit:
In [8]:
params = lmfit.Parameters()
params.add('a2', 0.5, min=0, max=1)
params.add('mu1', -1, min=-5, max=5)
params.add('mu2', 1, min=-5, max=5)
params.add('sig1', 1, min=1e-6)
params.add('sig2', 1, min=1e-6)
params.add('ax', expr='a2') # just a test for a derived parameter
Results = [lmfit.minimize(log_likelihood_lmfit, params, args=(di,),
nan_policy='omit', method='least_squares')
for di in d]
Fit results can be inspected with lmfit.fit_report() or
params.pretty_print():
In [9]:
print(lmfit.fit_report(Results[0]))
print()
Results[0].params.pretty_print()
[[Fit Statistics]]
# function evals = 137
# data points = 1
# variables = 6
chi-square = 3136682.002
reduced chi-square = -627336.400
Akaike info crit = 26.959
Bayesian info crit = 14.959
[[Variables]]
a2: 0.39327059 (init= 0.5)
mu1: -1.96272254 (init=-1)
mu2: 2.13070136 (init= 1)
sig1: 0.49895469 (init= 1)
sig2: 1.45925458 (init= 1)
ax: 0.39327059 == 'a2'
[[Correlations]] (unreported correlations are < 0.100)
Name Value Min Max Stderr Vary Expr
a2 0.3933 0 1 None True None
ax 0.3933 -inf inf None False a2
mu1 -1.963 -5 5 None True None
mu2 2.131 -5 5 None True None
sig1 0.499 1e-06 inf None True None
sig2 1.459 1e-06 inf None True None
This is good for peeking at the results. However, extracting these data from lmfit objects is quite a chore and requires good knowledge of lmfit objects structure.
pybroom helps in this task: it extracts data from fit results and returns familiar pandas DataFrame (in tidy format). Thanks to the tidy format these data can be much more easily manipulated, filtered and plotted.
Let’s use the glance and tidy functions:
In [10]:
dg = br.glance(Results)
dg.drop('message', 1).head()
Out[10]:
| method | num_params | num_data_points | chisqr | redchi | AIC | BIC | num_func_eval | success | key | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | least_squares | 6 | 1 | 3.136682e+06 | -627336.400468 | 26.958676 | 14.958676 | 137 | True | 0 |
| 1 | least_squares | 6 | 1 | 3.060677e+06 | -612135.352681 | 26.934147 | 14.934147 | 112 | True | 1 |
| 2 | least_squares | 6 | 1 | 3.260012e+06 | -652002.425980 | 26.997241 | 14.997241 | 141 | True | 2 |
| 3 | least_squares | 6 | 1 | 3.096436e+06 | -619287.111223 | 26.945762 | 14.945762 | 105 | True | 3 |
| 4 | least_squares | 6 | 1 | 3.097056e+06 | -619411.157122 | 26.945962 | 14.945962 | 145 | True | 4 |
In [11]:
dt = br.tidy(Results, var_names='dataset')
dt.query('dataset == 0')
Out[11]:
| name | value | min | max | vary | expr | stderr | init_value | dataset | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | a2 | 0.393271 | 0.000000 | 1.000000 | True | None | NaN | 0.000000 | 0 |
| 1 | ax | 0.393271 | -inf | inf | False | a2 | NaN | NaN | 0 |
| 2 | mu1 | -1.962723 | -5.000000 | 5.000000 | True | None | NaN | -0.201358 | 0 |
| 3 | mu2 | 2.130701 | -5.000000 | 5.000000 | True | None | NaN | 0.201358 | 0 |
| 4 | sig1 | 0.498955 | 0.000001 | inf | True | None | NaN | 1.732050 | 0 |
| 5 | sig2 | 1.459255 | 0.000001 | inf | True | None | NaN | 1.732050 | 0 |
Note that while glance returns one row per fit result, the tidy function return one row per fitted parameter.
We can query the value of one parameter (peak position) across the multiple datasets:
In [12]:
dt.query('name == "mu1"').head()
Out[12]:
| name | value | min | max | vary | expr | stderr | init_value | dataset | |
|---|---|---|---|---|---|---|---|---|---|
| 2 | mu1 | -1.962723 | -5.0 | 5.0 | True | None | NaN | -0.201358 | 0 |
| 8 | mu1 | -2.003590 | -5.0 | 5.0 | True | None | NaN | -0.201358 | 1 |
| 14 | mu1 | -1.950642 | -5.0 | 5.0 | True | None | NaN | -0.201358 | 2 |
| 20 | mu1 | -2.015723 | -5.0 | 5.0 | True | None | NaN | -0.201358 | 3 |
| 26 | mu1 | -2.009203 | -5.0 | 5.0 | True | None | NaN | -0.201358 | 4 |
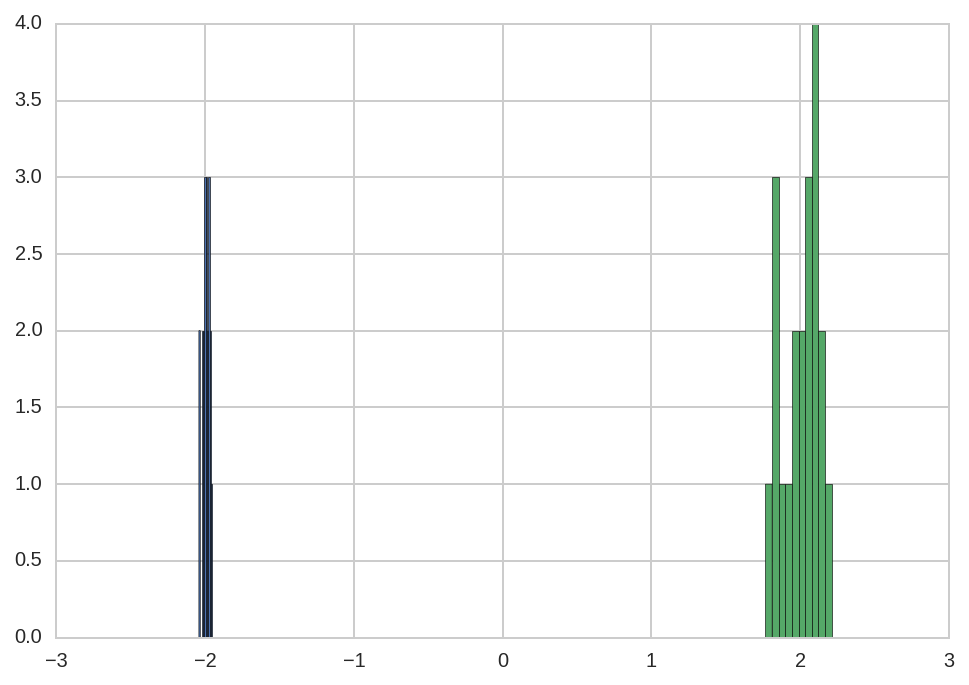
By computing the standard deviation of the peak positions:
In [13]:
dt.query('name == "mu1"')['value'].std()
Out[13]:
0.024538412206652392
In [14]:
dt.query('name == "mu2"')['value'].std()
Out[14]:
0.12309821388429468
we see that the estimation of mu1 as less error than the estimation
of mu2.
This difference can be also observed in the histogram of the fitted values:
In [15]:
dt.query('name == "mu1"')['value'].hist()
dt.query('name == "mu2"')['value'].hist(ax=plt.gca());
We can also use pybroom’s tidy_to_dict and dict_to_tidy functions to convert a set of fitted parameters to a dict (and vice-versa):
In [16]:
kwd_params = br.tidy_to_dict(dt.loc[dt['dataset'] == 0])
kwd_params
Out[16]:
{'a2': 0.39327059477457676,
'ax': 0.39327059477457676,
'mu1': -1.962722540352284,
'mu2': 2.1307013636579843,
'sig1': 0.49895469278155413,
'sig2': 1.4592545828997032}
In [17]:
br.dict_to_tidy(kwd_params)
Out[17]:
| name | value | |
|---|---|---|
| 0 | a2 | 0.393271 |
| 1 | ax | 0.393271 |
| 2 | mu1 | -1.962723 |
| 3 | mu2 | 2.130701 |
| 4 | sig1 | 0.498955 |
| 5 | sig2 | 1.459255 |
This conversion is useful to call a python functions passing argument values from a tidy DataFrame.
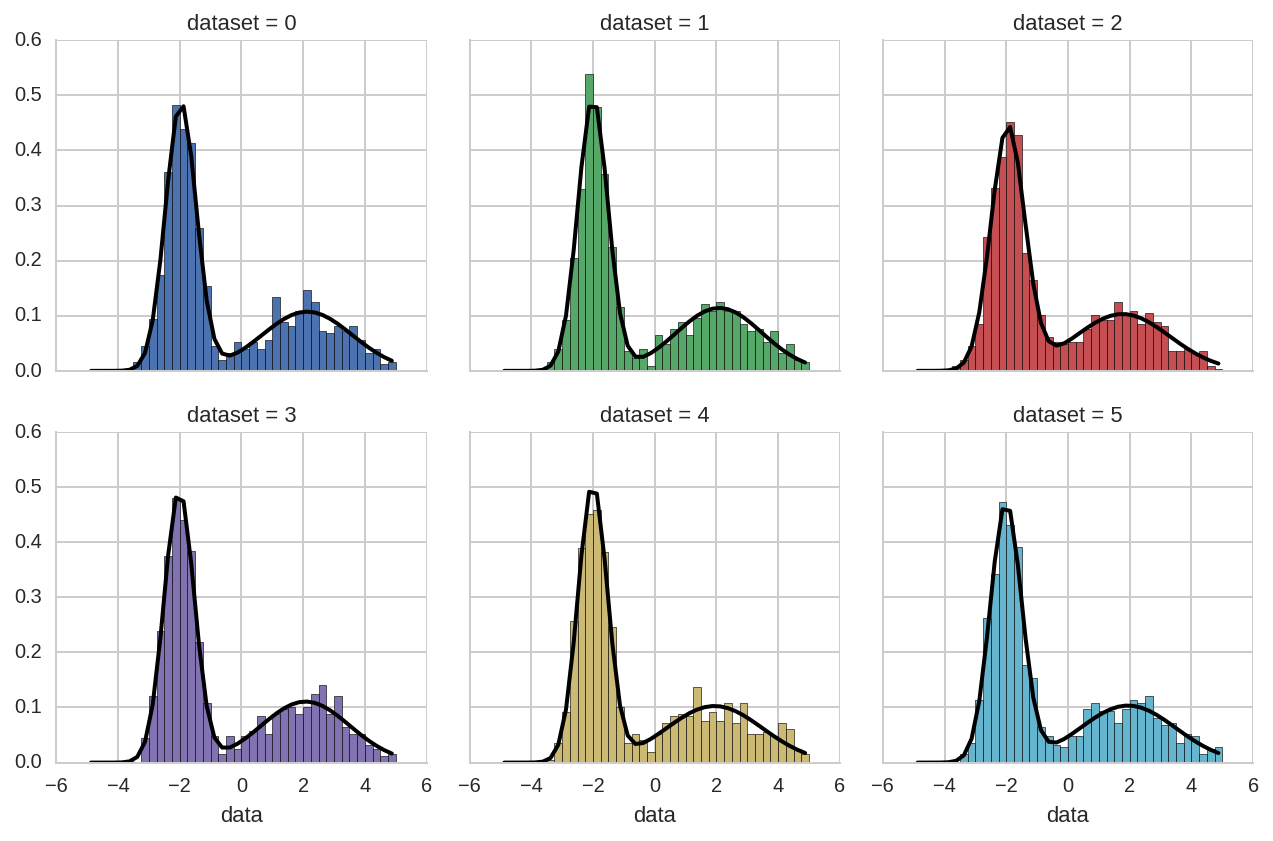
For example, here we use tidy_to_dict to easily plot the model
distribution:
In [18]:
bins = np.arange(-5, 5.01, 0.25)
x = bins[:-1] + 0.5*(bins[1] - bins[0])
grid = sns.FacetGrid(ds.query('dataset < 6'), col='dataset', hue='dataset', col_wrap=3)
grid.map(plt.hist, 'data', bins=bins, normed=True);
for i, ax in enumerate(grid.axes):
kw_pars = br.tidy_to_dict(dt.loc[dt.dataset == i], keys_exclude=['ax'])
y = model_pdf(x, **kw_pars)
ax.plot(x, y, lw=2, color='k')
Single-peak model¶
For the sake of the example we also fit the \(N\) datasets with a single Gaussian distribution:
In [19]:
def model_pdf1(x, mu, sig):
return normpdf(x, mu, sig)
def log_likelihood_lmfit1(params, x):
return -np.log(model_pdf1(x, **params.valuesdict())).sum()
In [20]:
params = lmfit.Parameters()
params.add('mu', 0, min=-5, max=5)
params.add('sig', 1, min=1e-6)
Results1 = [lmfit.minimize(log_likelihood_lmfit1, params, args=(di,),
nan_policy='omit', method='least_squares')
for di in d]
In [21]:
dg1 = br.glance(Results)
dg1.drop('message', 1).head()
Out[21]:
| method | num_params | num_data_points | chisqr | redchi | AIC | BIC | num_func_eval | success | key | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | least_squares | 6 | 1 | 3.136682e+06 | -627336.400468 | 26.958676 | 14.958676 | 137 | True | 0 |
| 1 | least_squares | 6 | 1 | 3.060677e+06 | -612135.352681 | 26.934147 | 14.934147 | 112 | True | 1 |
| 2 | least_squares | 6 | 1 | 3.260012e+06 | -652002.425980 | 26.997241 | 14.997241 | 141 | True | 2 |
| 3 | least_squares | 6 | 1 | 3.096436e+06 | -619287.111223 | 26.945762 | 14.945762 | 105 | True | 3 |
| 4 | least_squares | 6 | 1 | 3.097056e+06 | -619411.157122 | 26.945962 | 14.945962 | 145 | True | 4 |
In [22]:
dt1 = br.tidy(Results1, var_names='dataset')
dt1.query('dataset == 0')
Out[22]:
| name | value | min | max | vary | expr | stderr | init_value | dataset | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | mu | -0.352842 | -5.000000 | 5.000000 | True | NaN | NaN | 0.00000 | 0 |
| 1 | sig | 2.233056 | 0.000001 | inf | True | NaN | NaN | 1.73205 | 0 |
Augment?¶
Pybroom
augment
function extracts information that is the same size as the input
dataset, for example the array of residuals. In this case, however, we
performed a scalar minimization (the log-likelihood function returns a
scalar) and therefore the MinimizerResult object does not contain
any residual array or other data of the same size as the dataset.
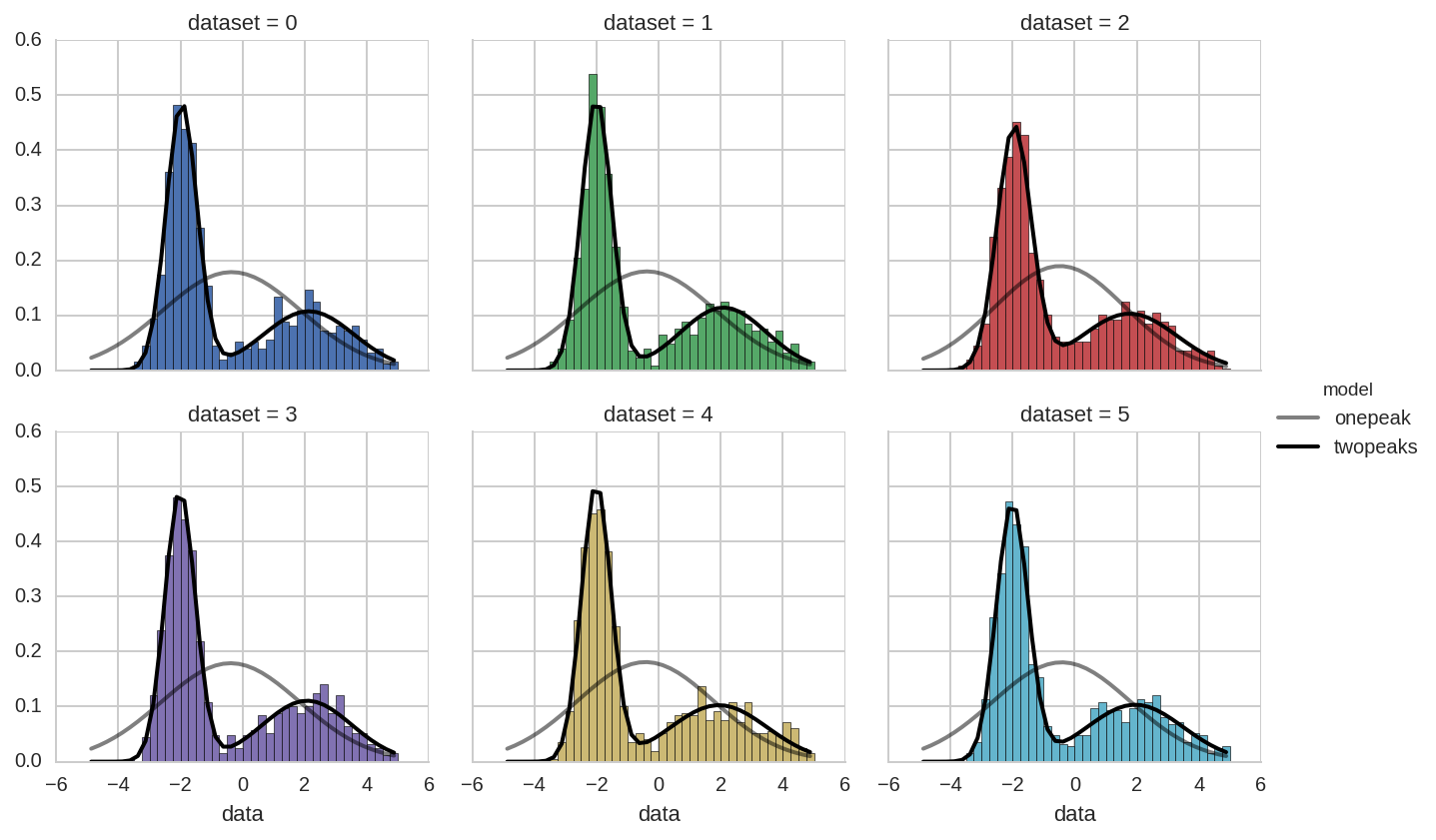
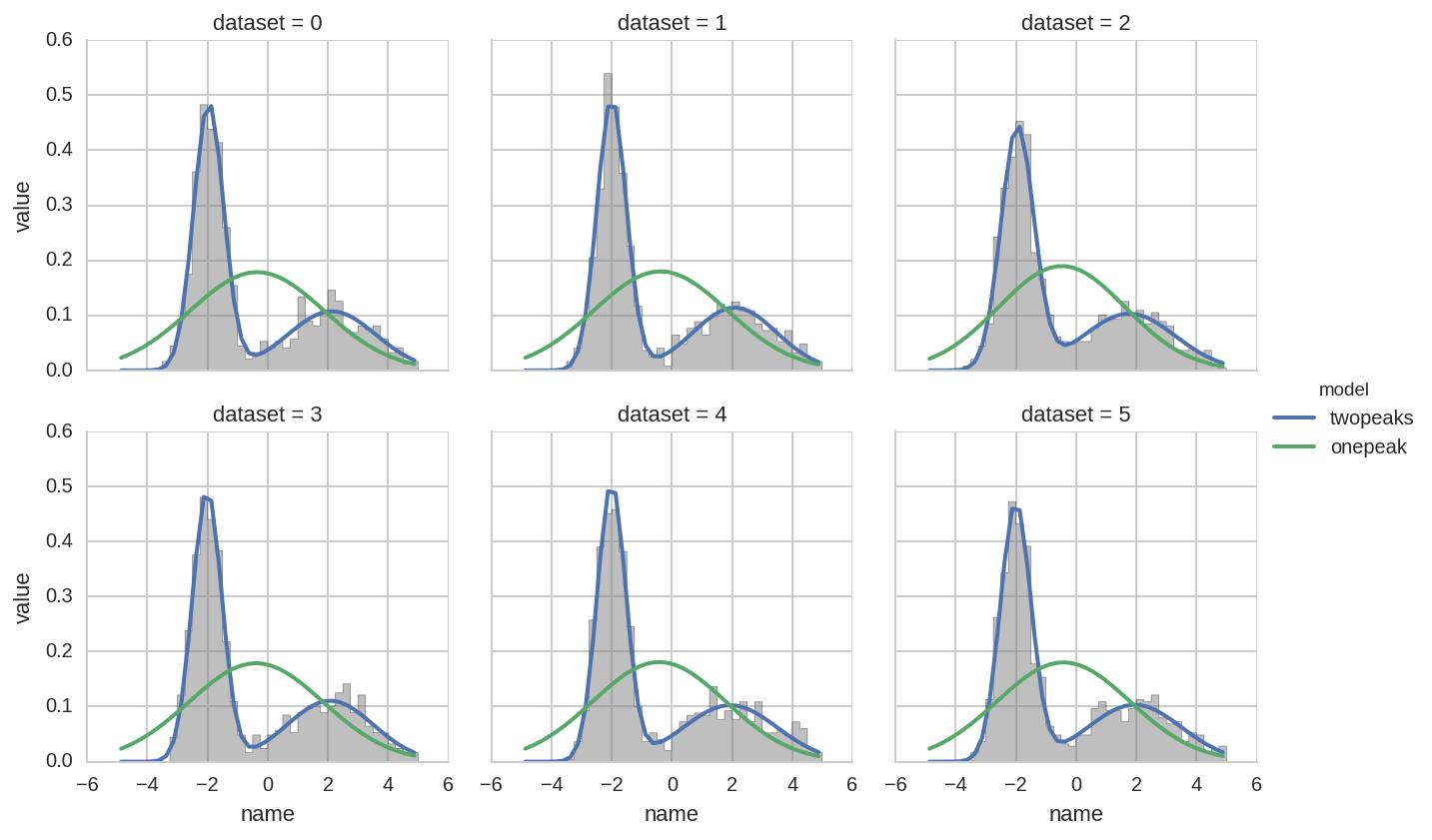
Comparing fit results¶
We will do instead a comparison of single and two-peaks distribution
using the results from the tidy function obtained in the previous
section.
We start with the following plot:
In [23]:
dt['model'] = 'twopeaks'
dt1['model'] = 'onepeak'
dt_tot = pd.concat([dt, dt1], ignore_index=True)
In [24]:
bins = np.arange(-5, 5.01, 0.25)
x = bins[:-1] + 0.5*(bins[1] - bins[0])
grid = sns.FacetGrid(ds.query('dataset < 6'), col='dataset', hue='dataset', col_wrap=3)
grid.map(plt.hist, 'data', bins=bins, normed=True);
for i, ax in enumerate(grid.axes):
kw_pars = br.tidy_to_dict(dt_tot.loc[(dt_tot.dataset == i) & (dt_tot.model == 'onepeak')])
y1 = model_pdf1(x, **kw_pars)
li1, = ax.plot(x, y1, lw=2, color='k', alpha=0.5)
kw_pars = br.tidy_to_dict(dt_tot.loc[(dt_tot.dataset == i) & (dt_tot.model == 'twopeaks')], keys_exclude=['ax'])
y = model_pdf(x, **kw_pars)
li, = ax.plot(x, y, lw=2, color='k')
grid.add_legend(legend_data=dict(onepeak=li1, twopeaks=li),
label_order=['onepeak', 'twopeaks'], title='model');
The problem is that FacetGrid only takes one DataFrame as input. In
the previous example we provide the DataFrame of “experimental” data
(ds) and use the .map method to plot histograms of the different
datasets. The fitted distributions, instead, are plotted manually in the
for loop.
We can invert the approach, and pass to FacetGrid the DataFrame of
fitted parameters (dt_tot), while leaving the simple histogram for
manual plotting. In this case we need to write an helper function
(_plot) that knows how to plot a distribution given a set of
parameter:
In [25]:
def _plot(names, values, x, label=None, color=None):
df = pd.concat([names, values], axis=1)
kw_pars = br.tidy_to_dict(df, keys_exclude=['ax'])
func = model_pdf1 if label == 'onepeak' else model_pdf
y = func(x, **kw_pars)
plt.plot(x, y, lw=2, color=color, label=label)
In [26]:
bins = np.arange(-5, 5.01, 0.25)
x = bins[:-1] + 0.5*(bins[1] - bins[0])
grid = sns.FacetGrid(dt_tot.query('dataset < 6'), col='dataset', hue='model', col_wrap=3)
grid.map(_plot, 'name', 'value', x=x)
grid.add_legend()
for i, ax in enumerate(grid.axes):
ax.hist(ds.query('dataset == %d' % i).data, bins=bins, histtype='stepfilled', normed=True,
color='gray', alpha=0.5);
For an even better (i.e. simpler) example of plots of fit results see pybroom-example-multi-datasets.